Why are eukaryotic mRNAs normally not polycistronic?
Let’s break it down:
Polycistronic mRNA: Imagine a single mRNA molecule containing instructions for multiple proteins. This is common in bacteria, where a single mRNA can code for several different proteins needed for a specific task.
Monocistronic mRNA: On the other hand, eukaryotic mRNAs are generally monocistronic, meaning each mRNA molecule only carries the instructions for one protein.
The reason for this difference lies in the way eukaryotic cells translate their genetic information. Eukaryotic mRNAs have a 5′ cap and a poly-A tail that act as signals for the ribosome, the protein-building machinery, to initiate and complete translation. Since each mRNA only has one starting point for translation, it only produces one protein.
Think of it this way: in bacteria, it’s like having a single cookbook with multiple recipes, all ready to be cooked. But in eukaryotes, each recipe gets its own cookbook, so the kitchen can focus on one dish at a time.
This difference in mRNA structure is linked to the way eukaryotic cells organize their genes. Eukaryotic genes are often spread out across the genome, and each gene has its own regulatory elements, allowing for precise control of gene expression. Having a dedicated mRNA for each gene ensures that the right proteins are made at the right time and in the right amounts.
So, even though eukaryotic mRNAs are generally shorter than bacterial mRNAs, the reason they’re not polycistronic is primarily due to the way the translation process is regulated. The cap and tail on eukaryotic mRNAs provide a distinct starting point for translation, ensuring that each mRNA produces only one protein. This allows for precise control over protein synthesis and helps maintain the complexity of eukaryotic cells.
Why is prokaryotic mRNA polycistronic while eukaryotic mRNA is monocistronic?
On the other hand, eukaryotic mRNA is monocistronic, meaning it only encodes for a single protein. This is because eukaryotes have a nucleus, which separates the transcription and translation processes. In eukaryotic cells, transcription takes place in the nucleus, and the resulting mRNA is then transported to the cytoplasm for translation. This separation allows for more complex regulation of gene expression in eukaryotes.
The difference between polycistronic and monocistronic mRNA stems from the organization of genes and the regulatory mechanisms involved in protein synthesis. In prokaryotes, genes involved in a particular metabolic pathway are often clustered together on the chromosome. This organization allows for coordinated expression of these genes, ensuring that all the proteins required for the pathway are produced simultaneously. The polycistronic mRNA allows for the translation of all these proteins from a single transcript, optimizing efficiency.
Eukaryotic genes, however, are often dispersed across the genome. This requires independent regulation of each gene. Monocistronic mRNA ensures that each gene is transcribed and translated independently, allowing for fine-tuning of gene expression to meet specific cellular needs.
Think of it this way: In prokaryotes, it’s like having a single cookbook that contains all the recipes for a multi-course meal, allowing you to prepare everything at once. In eukaryotes, it’s like having individual cookbooks for each dish, allowing you to prepare each dish independently.
Why is eukaryotic DNA monocistronic?
Eukaryotic mRNAs are typically monocistronic, meaning they each encode a single polypeptide. This is in contrast to prokaryotic mRNAs, which are often polycistronic, meaning they code for multiple polypeptides.
Why the difference? It comes down to how eukaryotic and prokaryotic cells regulate their gene expression.
Eukaryotic cells have a more complex system for gene regulation, with multiple steps involved in transcription and translation. This complexity demands a more organized approach, which is why eukaryotic mRNAs are monocistronic.
Imagine a chef preparing a multi-course meal. Each course requires specific ingredients and preparation. In this analogy, eukaryotic monocistronic mRNAs are like individual recipes, each focusing on creating a single dish (a polypeptide). This allows for precise control over the synthesis of each protein.
Prokaryotes, on the other hand, have a simpler system for gene regulation. They often need to quickly adapt to changes in their environment, which necessitates a more efficient way of producing proteins. Polycistronic mRNAs enable this efficiency by allowing multiple proteins to be produced from a single transcript, like a chef using a single recipe to make several dishes simultaneously.
In summary, the difference in the structure of eukaryotic and prokaryotic mRNAs is a reflection of the different strategies these organisms use to regulate gene expression. Eukaryotic cells prefer a more controlled and precise approach, while prokaryotes prioritize speed and efficiency.
Can eukaryotes have polycistronic genes?
Let’s break down what this means. Polycistronic mRNA refers to a single messenger RNA molecule that codes for multiple proteins. In contrast, monocistronic mRNA, the more common type in eukaryotes, carries the genetic code for just one protein. So, while eukaryotes can technically have polycistronic genes, they typically use a different system.
Here’s why this difference exists:
Translation in Prokaryotes: In prokaryotes, translation can begin even before transcription is finished. This means multiple ribosomes can attach to a single mRNA strand and start making proteins from different regions simultaneously. This is possible because prokaryotes lack a nucleus, allowing the processes of transcription and translation to happen in the same compartment.
Translation in Eukaryotes: Eukaryotic cells have a nucleus, separating the processes of transcription and translation. Transcription occurs in the nucleus, while translation occurs in the cytoplasm. This means that the mRNA must be processed and transported out of the nucleus before translation can occur.
Eukaryotic genes are also organized differently. They often have introns, non-coding sequences within the gene, that need to be removed before translation. This processing step further complicates the production of multiple proteins from a single mRNA.
While polycistronic mRNAs are rare in eukaryotes, there are some exceptions. For example, some viruses that infect eukaryotic cells use polycistronic mRNAs to produce multiple proteins from a single transcript. Additionally, some eukaryotes have been found to use polycistronic mRNAs for specific purposes, such as in the regulation of gene expression. While less common, polycistronic arrangements demonstrate the flexibility and adaptability of gene expression in eukaryotes.
Can mRNA be Polycistronic?
Polycistronic mRNA refers to a single messenger RNA molecule that carries the genetic code for multiple proteins. Think of it like a multi-purpose recipe, where one instruction set leads to the creation of several different dishes. This is common in bacteria (prokaryotes) and chloroplasts, tiny organelles found in plant cells.
Why is this important? Well, it’s all about efficiency. By encoding multiple proteins on a single mRNA molecule, bacteria and chloroplasts can save energy and resources. Imagine trying to make a cake and having to follow a separate recipe for each ingredient! Polycistronic mRNA is like having one recipe that covers everything, from the batter to the icing.
But here’s the catch: eukaryotic cells (like those found in animals and plants) don’t usually use polycistronic mRNAs. Instead, they typically have one mRNA molecule for each protein. This difference is rooted in the way eukaryotic cells organize their genes. Eukaryotic genes often have “non-coding” regions called introns that need to be removed before the mRNA can be translated into protein. This complex process makes it difficult to have multiple protein-coding sequences on a single mRNA molecule.
In summary, polycistronic mRNA is an efficient way for bacteria and chloroplasts to produce multiple proteins from a single genetic message. While eukaryotic cells typically use separate mRNAs for each protein, the world of mRNA is incredibly diverse and holds many secrets yet to be discovered!
Why is mRNA spliced in eukaryotes but not in prokaryotes?
You might be wondering why mRNA undergoes splicing in eukaryotic cells but not in prokaryotic cells. The answer lies in the structure of the mRNA itself.
Eukaryotic mRNA contains exons (coding sequences) and introns (non-coding sequences). Think of it like a recipe: the exons are the actual instructions for making a dish, while the introns are just notes or comments that don’t affect the final outcome. Before the mRNA can be used to make proteins, the introns need to be removed, and the exons need to be spliced together. This process is called mRNA splicing.
Prokaryotic mRNA, on the other hand, doesn’t have introns. It’s like a recipe with only instructions, no extra notes. This means that prokaryotic mRNA can be translated into proteins directly without any splicing.
Why the difference?
The reason for this difference lies in the way eukaryotic and prokaryotic cells organize their genes. In eukaryotes, genes are often broken up into smaller pieces (exons) with intervening non-coding regions (introns). This allows for more complex gene regulation and protein diversity. Prokaryotes, on the other hand, generally have their genes in continuous stretches, making their mRNA simpler.
What happens during splicing?
The splicing process is complex and involves a number of specialized proteins called spliceosomes. The spliceosome recognizes the boundaries between exons and introns and cuts out the introns, then joins the exons together. This ensures that only the coding sequences are used to build proteins.
A few extra things to note:
* Splicing isn’t just about removing introns; it can also lead to alternative splicing, where different combinations of exons can be spliced together. This allows a single gene to produce multiple different proteins, further increasing the complexity of eukaryotic gene expression.
* The splicing process is crucial for the correct functioning of eukaryotic cells. Errors in splicing can lead to the production of non-functional proteins, which can cause a variety of diseases.
In summary, splicing is a unique feature of eukaryotic mRNA that allows for more complex gene regulation and protein diversity. Prokaryotic mRNA does not undergo splicing because it is a simpler system where genes are typically continuous.
Is mRNA always monocistronic?
Eukaryotic mRNAs are monocistronic. This means that each mRNA molecule carries the genetic code for one protein. Prokaryotic mRNAs, on the other hand, can be monocistronic or polycistronic, which means they encode for one or multiple proteins, respectively.
Bacteriophages, which are viruses that infect bacteria, can also have monocistronic or polycistronic mRNAs, similar to their prokaryotic hosts.
So, why are eukaryotic mRNAs always monocistronic?
This is due to the intricate way eukaryotic cells translate their genetic code. Eukaryotic translation starts at a specific sequence called the 5′ cap, and proceeds along the mRNA until it encounters a stop codon. This process creates a single polypeptide chain, ensuring that each mRNA molecule only encodes a single protein.
In contrast, prokaryotic translation is more flexible. Prokaryotes use a system called operons, where multiple genes are clustered together under the control of a single promoter. This allows for the coordinated expression of multiple proteins, which is crucial for rapid responses to environmental changes.
Here’s a simplified breakdown:
Eukaryotes: Each mRNA molecule is like a train with a single passenger (protein).
Prokaryotes: Some mRNAs are like trains with one passenger, while others are like trains with multiple passengers.
Understanding these differences is essential for comprehending the diverse ways organisms express their genetic information. This knowledge is crucial for developing new therapeutic strategies, such as mRNA vaccines, which rely on manipulating the translation process.
Why might it be problematic to use a eukaryotic translation system with polycistronic mRNA?
Eukaryotic cells typically process monocistronic mRNAs, which means each mRNA molecule encodes for only one polypeptide. This is different from prokaryotic cells, which often use polycistronic mRNAs that can encode for multiple proteins.
So, why might using polycistronic mRNA in a eukaryotic system be a challenge? It’s because the eukaryotic translation machinery is optimized for recognizing and translating single protein-coding sequences. This is due to several key features:
Ribosome Binding Sites: In eukaryotic cells, each mRNA molecule usually has one ribosome binding site, also known as a 5′ cap, where the ribosome attaches to begin translation. In polycistronic mRNAs, multiple proteins are encoded in a single mRNA, so there would be multiple ribosome binding sites. This can create confusion for the translation machinery, leading to inefficient or incorrect translation of the different proteins.
Translation Initiation Factors: Eukaryotic cells rely on specific translation initiation factors (eIFs) to help ribosomes recognize the 5′ cap and start translating the mRNA. These factors wouldn’t necessarily be able to efficiently handle multiple ribosome binding sites on a polycistronic mRNA.
mRNA Processing: Eukaryotic cells modify their mRNAs with a 5′ cap and a poly(A) tail. These modifications play crucial roles in stabilizing the mRNA, protecting it from degradation, and facilitating its transport to the cytoplasm for translation. In a polycistronic mRNA, these modifications would need to be properly positioned and coordinated for each individual protein-coding sequence, which could add complexity to the processing and translation process.
In summary, eukaryotic cells are designed to handle monocistronic mRNAs efficiently. While it’s possible to engineer eukaryotic systems to express polycistronic mRNAs, it often requires significant optimization to ensure that all the encoded proteins are translated correctly and efficiently.
Are operons only in prokaryotes?
Operons are groups of genes that are transcribed together as a single unit. This allows for the coordinated regulation of gene expression, which is essential for the efficient functioning of prokaryotic cells. While operons are less common in eukaryotes, there are a few key differences that explain their prevalence in prokaryotes:
Gene Regulation: Prokaryotic cells often need to rapidly respond to changes in their environment, and operons allow for the coordinated regulation of multiple genes involved in a particular metabolic pathway.
Transcriptional Efficiency: Transcribing multiple genes as a single unit is more efficient than transcribing them individually. This is especially important for prokaryotic cells that have a smaller genome size and limited resources.
Genome Organization: Prokaryotic genomes are generally smaller and more compact than eukaryotic genomes. This makes it easier to organize genes into operons.
While eukaryotes generally use more complex regulatory mechanisms, the presence of operons in some eukaryotes suggests that these structures might have been more prevalent in the past. Additionally, the identification of operons in diverse eukaryotic species highlights the evolutionary conservation of this gene regulatory strategy.
Why is eukaryotic DNA linear?
Think of it this way: imagine you have a giant library full of books. To organize it, you wouldn’t just throw all the books in a single huge pile. You’d divide the books into manageable sections, using shelves and labels to keep things neat and accessible. In a similar way, linear chromosomes act like the shelves in our cells, breaking down the massive amount of DNA into organized chunks. This makes it easier for the cellular machinery to find the right genes, copy them when needed, and keep everything in order.
The ability to efficiently manage large genomes likely played a crucial role in the evolution of complex eukaryotic organisms. As lifeforms became more intricate, their genetic instructions grew more elaborate, demanding a system capable of handling the increased complexity. Linear chromosomes, with their organized structure and ability to handle large amounts of information, may have provided the evolutionary advantage needed for eukaryotes to thrive.
See more here: Why Is Prokaryotic Mrna Polycistronic While Eukaryotic Mrna Is Monocistronic? | Why Are Eukaryotic Mrnas Not Polycistronic
Are eukaryotic mRNAs polycistronic?
A gene family in insects has been found to produce several short peptides, some as small as 11 amino acids, each with its own start and stop codon. This means that a single mRNA molecule can encode for multiple proteins, defying the traditional notion of monocistronic mRNA in eukaryotes.
Let’s delve deeper into why this discovery is so significant. In eukaryotes, the process of translation, where mRNA is used to create proteins, usually starts at a specific start codon (AUG) and ends at a stop codon (UAG, UAA, or UGA). This ensures that only one protein is produced from each mRNA molecule.
However, the newly discovered gene family in insects breaks this rule. The mRNA molecule contains multiple start and stop codons, allowing the production of multiple short peptides from a single transcript. This opens up a whole new world of possibilities for how proteins are made and regulated in these insects.
This discovery also suggests that there might be other instances of polycistronic mRNA in eukaryotes, which could potentially have significant implications for our understanding of gene expression and regulation. It’s a reminder that the world of genetics is constantly evolving and there’s still much to learn about the complex mechanisms that govern life.
Do eukaryotes have polycistronic gene structures?
Let’s break down why eukaryotic mRNAs are typically monocistronic and how viruses exploit this system.
Eukaryotic mRNA Structure: Eukaryotic mRNAs have a 5′ cap and a poly(A) tail. The 5′ cap acts as a recognition signal for the ribosome, guiding it to the start codon. The poly(A) tail helps to stabilize the mRNA and is important for its export from the nucleus. Each mRNA molecule also has a coding region, which contains the genetic information for the protein. In eukaryotes, each coding region typically corresponds to one protein product. This is because eukaryotic mRNAs are often translated in a monocistronic fashion, meaning each mRNA molecule produces only one protein.
Polycistronic mRNA and Viruses: Now, let’s talk about polycistronic mRNAs. These mRNAs contain multiple open reading frames (ORFs), each of which codes for a different protein. In eukaryotes, polycistronic mRNAs are relatively rare, primarily found in viruses. Viruses often exploit this strategy to efficiently express multiple proteins needed for their replication and propagation.
How Viruses Deal with Polycistronic mRNA: The presence of polycistronic mRNAs in viruses necessitates a mechanism for the ribosome to recognize and translate multiple ORFs within a single mRNA molecule. Viruses have evolved clever strategies to achieve this. For instance, some viruses employ internal ribosome entry sites (IRESs) within their mRNAs. These IRESs are specialized sequences that allow the ribosome to bind to the mRNA and initiate translation at a point other than the traditional 5′ cap. By using IRESs, viruses can ensure that multiple ORFs within their polycistronic mRNAs are translated.
In summary, while eukaryotic mRNAs are typically monocistronic, the discovery of polycistronic mRNAs in viruses demonstrates the adaptability of the eukaryotic translation machinery. Viruses have ingeniously evolved ways to translate multiple proteins from a single mRNA molecule, highlighting the remarkable flexibility of this fundamental cellular process.
What is a polycistronic mRNA?
Polycistronic mRNA describes a single messenger RNA molecule that encodes multiple proteins. Think of it like a multi-pack of instructions. This is in contrast to monocistronic mRNA, which carries instructions for only one protein.
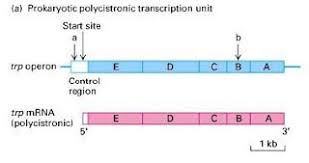
Polycistronic mRNAs are often called operons because they are controlled by a single promoter and terminator. This means that the expression of all the genes in the operon is regulated together.
So, how does it work? Well, a polycistronic mRNA is transcribed from a single gene but contains multiple start and stop codons. These codons tell the ribosome when to begin and end translation for each protein.
Here’s an example to illustrate: imagine you have a set of instructions for building a car. This set of instructions might include instructions for building the engine, the wheels, and the body. This would be similar to a polycistronic mRNA, where one message contains multiple sets of instructions (for different proteins).
Polycistronic mRNAs are primarily found in prokaryotes, like bacteria. This is because prokaryotes are simpler organisms and can benefit from the efficient use of resources.
On the other hand, eukaryotes, like humans and animals, have evolved to use monocistronic mRNAs. This means that each protein has its own separate mRNA molecule. While this might seem less efficient, it offers more flexibility and control over gene expression in complex organisms.
Now, let’s dive a little deeper into the significance of polycistronic mRNAs:
Efficient resource use: Since polycistronic mRNAs encode multiple proteins, they help prokaryotes save energy and resources. It’s like having one instruction manual for multiple tasks, reducing the need for individual manuals.
Coordinated gene expression: The operon structure allows for coordinated regulation of related genes. If a bacterium needs to break down a particular sugar, for example, it can turn on the expression of all the genes necessary for this process simultaneously by activating the operon. This ensures that the bacterium can quickly and efficiently respond to changes in its environment.
Flexibility: Operons can be regulated by various factors like repressors and activators, allowing for flexibility in gene expression.
In conclusion, polycistronic mRNAs are unique to prokaryotes and are important for their efficient use of resources and coordinated gene expression. Understanding these fascinating molecules provides insights into the diverse strategies employed by organisms to regulate their gene expression and adapt to their environments.
How eukaryotic mRNAs are modified?
After splicing, the mature mRNA is transported from the nucleus, where it was transcribed, to the cytoplasm, where protein synthesis takes place. This journey is essential for the efficient production of proteins.
But splicing isn’t the only modification that occurs. There are other key modifications that help the mRNA molecule perform its job:
5′ capping: This is like adding a protective cap to the beginning of the mRNA molecule. The 5′ cap helps protect the mRNA from degradation and also helps the ribosome, the protein-making machinery, bind to the mRNA.
3′ polyadenylation: This is like adding a tail to the end of the mRNA molecule. The poly-A tail helps stabilize the mRNA and also helps it get transported from the nucleus to the cytoplasm.
These modifications work together to ensure that the mRNA is stable, protected, and can be efficiently translated into a protein.
See more new information: bmxracingthailand.com
Why Are Eukaryotic Mrnas Not Polycistronic?
Okay, let’s talk about why eukaryotic mRNAs aren’t polycistronic. It’s actually a pretty important difference between prokaryotes and eukaryotes.
So, you might be asking yourself, “What’s a polycistronic mRNA?” Well, it’s a messenger RNA molecule that encodes for multiple proteins, all from a single transcript. Think of it like a multi-pack of cookies. One package contains multiple cookies, just like one polycistronic mRNA can hold the code for multiple proteins.
On the other hand, eukaryotic mRNAs are monocistronic. This means they only carry the genetic code for one protein. It’s like a single-serving pack of cookies – just one cookie per packet.
Why the Difference?
The main reason for this difference lies in how prokaryotes and eukaryotes organize their genes. Prokaryotes have operons, which are groups of genes that are transcribed together as a single unit. These operons are controlled by a single promoter and produce a single polycistronic mRNA.
Example: Think of the lac operon. This operon contains the genes for lactose metabolism. These genes are transcribed together, resulting in a single mRNA that carries the instructions for all the proteins needed for lactose breakdown.
Eukaryotes, however, are much more complex. Their genes are often scattered throughout their genome, and they need more regulation. That’s why they don’t use operons. Instead, each gene is transcribed independently, resulting in a monocistronic mRNA.
Translation and Ribosomes:
There’s another key factor involved – ribosomes. In prokaryotes, ribosomes can bind to mRNA and start translating proteins immediately after transcription. This is because transcription and translation happen in the same place – the cytoplasm.
But in eukaryotes, things are different. Transcription happens in the nucleus, and translation happens in the cytoplasm. So, the mRNA needs to be processed and transported out of the nucleus before translation can begin.
Translation Initiation: The Key Difference
The real difference between prokaryotic and eukaryotic translation initiation lies in how the ribosomes recognize the start codon.
Prokaryotes: The ribosome directly binds to the Shine-Dalgarno sequence located upstream of the start codon. This sequence is a key signal for the ribosome to recognize the start of a protein-coding region.
Eukaryotes: Ribosomes bind to the 5′ cap of the mRNA. They then scan along the mRNA until they find the start codon (AUG). This is where translation begins.
The 5′ cap is like a signal flag telling the ribosome where to start. This system allows eukaryotic cells to control translation initiation for each mRNA individually, ensuring that only one protein is made per mRNA.
So, in a nutshell:
Prokaryotes: Polycistronic mRNAs, operons, and ribosome binding directly to the Shine-Dalgarno sequence.
Eukaryotes: Monocistronic mRNAs, individual gene transcription, and ribosome binding to the 5′ cap.
Why is this important?
This difference in mRNA structure and translation initiation allows eukaryotes to control protein expression with greater precision. This is essential for complex multicellular organisms like ourselves.
Imagine if our cells couldn’t control which proteins were made and when! It would be a chaotic mess. Thanks to monocistronic mRNAs, we can keep things organized and ensure our cells function properly.
Let’s Summarize
Here’s a table summarizing the key differences between prokaryotic and eukaryotic mRNAs:
| Feature | Prokaryotic mRNA | Eukaryotic mRNA |
|—|—|—|
| Cistronic Nature | Polycistronic | Monocistronic |
| Gene Organization | Operons | Individual genes |
| Translation Initiation | Shine-Dalgarno sequence | 5′ cap |
FAQs
Q: Can eukaryotic mRNAs ever be polycistronic?
A: While it’s rare, there are some exceptions! In certain cases, viral mRNAs can be polycistronic. Viruses can sometimes hijack the host cell’s machinery and create their own unique mRNA structures.
Q: Why is the Shine-Dalgarno sequence important in prokaryotes?
A: The Shine-Dalgarno sequence is a ribosome-binding site. It helps guide the ribosome to the correct position on the mRNA so that translation can begin at the correct start codon.
Q: How does the 5′ cap affect translation initiation?
A: The 5′ cap acts as a recognition signal for the ribosome. It helps the ribosome bind to the mRNA and start scanning for the start codon.
Q: What are the benefits of monocistronic mRNAs in eukaryotes?
A: Monocistronic mRNAs allow for greater control over gene expression. This is crucial for complex organisms like humans, where different cells need to express different sets of proteins.
Q: Is the difference in mRNA structure between prokaryotes and eukaryotes significant?
A: Absolutely! This difference reflects the fundamental differences in gene regulation and protein synthesis between these two domains of life. It highlights the amazing diversity and complexity of life on Earth.
Understanding the difference between prokaryotic and eukaryotic mRNAs is crucial for comprehending the intricacies of gene expression and its vital role in all living organisms. It’s a fascinating area of biology that continues to unravel new discoveries and deepen our understanding of the molecular mechanisms that govern life itself.
RNA Processing and Turnover – The Cell – NCBI
The 3′ end of most eukaryotic mRNAs is defined not by termination of transcription, but by cleavage of the primary transcript and addition of a National Center for Biotechnology Information
Mammalian polycistronic mRNAs and disease – PMC – National
For viral and eukaryotic mRNAs, highly-structured IRES elements can form complexes with ribosomes together with a subset of the canonical eIFs and other RNA binding National Center for Biotechnology Information
Regulation of translation via mRNA structure in prokaryotes and …
The polycistronic structure of mRNAs is an important aspect of translational control in prokaryotes, but polycistronic mRNAs are not usable (and usually not PubMed
From Birth to Death: The Complex Lives of Eukaryotic mRNAs
In eukaryotes, mRNAs are first synthesized in the nucleus as pre-mRNAs that are subject to 5′-end capping, splicing, 3′-end cleavage, and polyadenylation. Once Science | AAAS
Roles of mRNA poly (A) tails in regulation of eukaryotic gene …
Poly (A) tails are present on almost every eukaryotic mRNA, with the only known exception being some mammalian histone transcripts. Poly (A) tails are added co Nature
what is poly… – Reasons &… | Learn Science at Scitable – Nature
All eukaryotic mRNAs are monocistronic. However, some prokaryotic mRNAs are monocistronic and others are polycistronic. Similar to prokaryotic mRNAs, bacteriophage Nature
All things must pass: contrasts and commonalities in eukaryotic
Despite its universal importance for controlling gene expression, mRNA degradation was initially thought to occur by disparate mechanisms in eukaryotes and Nature
Mammalian Polycistronic mRNAs and Disease:
In this review, we address the gradual recognition that a growing number of polycistronic genes, originally discovered in viruses, are being identified within the mammalian genome, and that these may Cell Press
A universal strategy for regulating mRNA translation in
One key characteristic of prokaryotic mRNAs is that they can be polycistronic. A polycistronic mRNA contains two or more cistrons, each of which can National Center for Biotechnology Information
Mcat Flashcard: Polycistronic Mrna
Differences In Translation Between Prokaryotes And Eukaryotes | Mcat | Khan Academy
Difference Between Polycistronic And Monocistronic Mrna | Rapid Fire Q.9
Polycistronic Mrna \U0026 Monocistronic Mrna With Dr. Sohail Jamil Qureshi L Mdcat Biology L Ap Biology
Transcription (Dna To Mrna)
Prokaryotic Vs. Eukaryotic Cells (Updated)
What Polycistronic Messanger Rna Means
Assertion (A) Transcription Unit Is Often Monocistronic In Eukaryotes And Polycistronic In Prokaryot
Comparing Mrna Transcription Processes Of Eukaryotes And Bacteria (Bios 041)
Translation Initiation In Eukaryotes | Eukaryotic Translation Lecture 1
Link to this article: why are eukaryotic mrnas not polycistronic.
See more articles in the same category here: bmxracingthailand.com/what
